With newly developed antisense oligonucleotide (ASO) molecules, early efficacy and toxicity assessment is crucial to prevent costly failures in later stages. But, what are the assays that will give you the best predictions to advance your drug candidate with confidence?
Here you can find 5 key steps you need to consider to successfully develop an ASO in vitro screening assay. Read till the end to discover the extra-advantage of designing a physiologically relevant assay that can save you time.
Background
Antisense oligonucleotides (ASOs) are a short, synthetic nucleic acid molecule that can bind to specific RNA sequences and can thereby modulate gene expression. This modulation can happen either through inducing degradation of the complimentary sequence, blocking translation or modulating splicing.
This trait can be incredibly valuable in the treatment of genetic disorders and hereditable diseases by either reducing target RNA transcript levels or restoring protein function.
Actually, some ASO therapeutics already made it to the market: Vitravene (Cytomegalovirus (CMV) retinitis)1, Kynamro (Familial Hypercholesterolemia)2, Tegsedi (TTR Polyneuropathy)3 and Waylivra (Familial Chylomicronemia Syndrome)4.
Nonetheless, safety concerns due to off target effects and limited efficacy are obstacles that need to be overcome, to deliver on the potential of ASOs as an effective therapeutic.
To enhance the likelihood of achieving late-stage success, early adoption of relevant cell-based in vitro screening assays can significantly influence subsequent outcomes.
Delve into 5 essential considerations to design relevant cell-based in vitro assays that empower you to obtain more accurate predictions regarding ASO efficacy and safety.
How to design a successful cell-based in vitro assay for ASO screening
1. Choosing a cell model relevant to your target disease
For the development of an in vitro assay, the basis lies on the selection of the right cell model. In principle, any cell line can be used to develop an assay, but you should look for the one that best fits your research questions and goals.
Each cell model has different (dis)advantages which you must know in advance to be able to get a balance between its physiological relevance, genetic stability, availability, reproducibility, scalability and cost.
One of the most common models used are immortalized cells. The advantage of an immortalized cell is, as the name suggests, the unlimited proliferative capacity.
However, they are not genetically stable and lack the capacity to replicate important cell functions or disease phenotypes. This considerably limits the options of getting accurate predictions for efficacy and toxicity.
Another option is to use primary human cells, which offer a high physiological relevance and enable researchers to mimic a diseased condition in vitro.
The disadvantage of this model is the limited proliferation capacity which hinders their application in primary screenings where large cell batches are required. Primary cells also present inter-donor variability, reducing the robustness of the assay, especially in the context of high-throughput screening.
Overall, primary human cells can be a valuable addition for validation studies where the need for scalability is much lower.
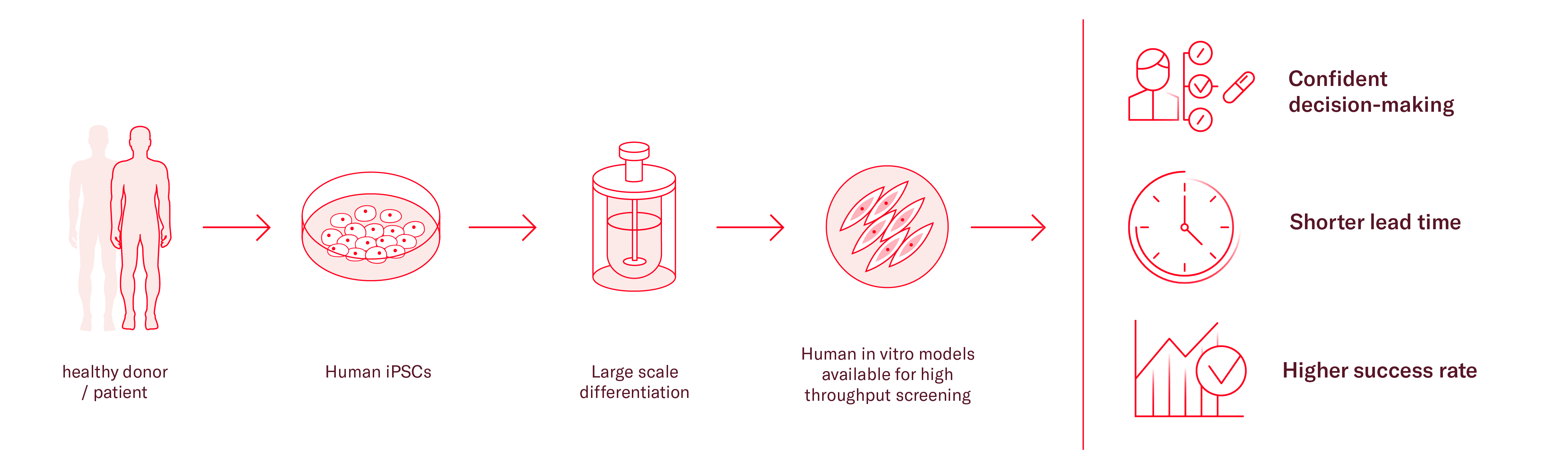
As an alternative, induced pluripotent stem cells (iPSCs) can be a useful tool in many cases, especially when investigating diseases where primary material is difficult to obtain, such as heart and brain related diseases. Availability of primary cells for those tissues is particularly limited, but iPSCs can be differentiated into neural and cardiac cells, as well as any body cell type.
Additionally, iPSCs have unlimited proliferation capacity and genetically stable. They can be patient-specific and recapitulate main disease phenotypes in vitro. To further enhance the relevance of the model and its translatability, different cell types can be cocultured to build more representative environments.
With the right expertise, iPSCs can be differentiated at a large scale producing one single source of cells for the full screening cascade, considerably reducing the variability of your results.
As there is no perfect model, the downside of iPSCs is the required technical expertise to develop robust differentiation procedures, produce large batches and model disease phenotypes in the context of miniaturized assays. Therefore, many researchers partner with expert companies to accelerate pipelines and save resources.
2. Selecting a relevant readout technology
There are three options you can consider to quantify the changes in the expression of your target.
Firstly, based on the abundance of transcript, you can choose either qPCR or ddPCR, selecting the latter when the abundance is low. Quantification of the RNA allows for the direct measurement of target engagement and is often the method of choice for many ASO screens.
The assay used to quantify the target mRNA needs to be optimized by adjusting the various parameters, such as thermocycling conditions, primer and probes designs and RNA quality and quantity, among others.
At this point, it is important to identify and evaluate at least two to three housekeeping genes (controls to normalize gene expression) prior to deciding on a specific one for your assay.
By systematically optimizing these parameters, you can enhance the sensitivity, specificity, and reliability of qPCR assays for quantifying target nucleic acid sequences.
Another option for quantification is high-content imaging (HCI) where either the RNA species can be visualized by fluorescence probes (FISH) or the target protein levels - end-product of your target gene – can be quantified.
However, this technique is only applicable when there are specific antibodies available for your target. If that’s the case, you might consider HCI instead of PCR for the following advantages:
- Quantifying protein expression with HCI allows the evaluation of the effectiveness of ASO treatment in achieving the desired alterations in protein expression.
- HCI can be multiplexed with additional markers for cell health or disease cellular pathways, providing a more holistic understanding of mechanism of action and toxicity.
3. Understanding the expression profile of your target gene
After the selection of the target and cell model system, the cellular models need to be cultured under physiological conditions and the kinetics of the target gene established.
This will ensure that the cells express the target mRNA at detectable and robust levels and the readouts are performed at a time point when the target levels (whether that be mRNA or protein) have stabilized.
Next step is to ensure that the cellular model is amenable to oligonucleotide transfection or delivery.
4. Selecting a method for ASO delivery
The most prevalent method is gymnotic uptake, where the ASOs enter the cell without the use of transfection reagents just following the principle of passive diffusion through the membrane.
This technique is highly dependent on properties of the ASO molecules, as well as the target cell and its membrane permeability. Therefore, you may need to optimized the delivery method to achieve efficient oligonucleotide uptake and intracellular delivery.
If the gymnotic method does not deliver enough ASO, transfection reagents or nucleofection can be considered. This technique is very suitable for delivering nucleic acids across the cell membrane, but it is vital to optimize it to prevent cytotoxicity while achieving modulation of the target gene.
For optimization purposes, it is recommend to perform qPCR of the target gene with positive control ASOs to ensure that most efficient delivery method of the ASOs is selected.
5. Optimize assay conditions
The screening protocol is further optimized to maximize sensitivity, specificity, and reproducibility by determining the appropriate:
- Oligonucleotide concentrations – normally determined by performing an 8 or 10 point concentration response curve (CRC).
- Duration of the ASO exposure – different study designs can be applied, but frequently the main decision is between an acute treatment (3 – 5 day exposure) or a chronic treatment (with multiple ASO additions through media refreshments).
- Overall assay conditions - performing pilot experiments that would mimic the workflow of the primary screen.
It is advised to identify both positive and negative control ASOs as it helps with the assay optimization and benchmarking the performance of the assay.
To effectively and robustly screen large libraries, it is recommended to automate the different steps of the screening process: e.g. cell seeding, media refreshes, ASO addition, assay readout, data analysis, etc. This helps you make a more confident selection of the hits before moving into potency determination or functional studies.
The advantage of a physiologically relevant design - save time by running complementary assays simultaneously
Once your assay is designed your next question is how to make accurate predictions on ASO in vivo efficacy and toxicity.
In both scenarios, the use of clinically relevant readouts is paramount for generating accurate predictions and advancing your therapeutic to next stages. It is important to recognize that the range of available readouts is heavily influenced by the choice of cell model made in step 1.
When selecting a relevant human cell model, there is the additional advantage of using the same system for complementary assays, saving time and resources.

For example, you can consider adding early toxicity readouts where ASOs are added to neuronal or cardiac cells to predict potential in vivo cardiotoxicity or neuronal liabilities. The toxicity of the ASO can be estimated through changes in calcium signaling or cellular metabolism, among other readouts.
Another range of assays that can help you select the most promising ASOs earlier, focuses on evaluating efficacy to rescue the disease phenotype. This can determine if the knockout or knock-down of the target gene is actually reversing or reducing important disease hallmarks.
Once more, this is only relevant if the cellular model can recapitulate disease phenotypes.
Through these assays, you can evaluate the direct effect of the therapeutic candidate on the disease phenotype.
Conclusions
When developing an ASO for therapeutic applications, early identification and validation of safety and efficacy are pivotal for success in both preclinical and clinical studies.
You must be aware of the profoundly influence that the cell model, readout and assay setup of choice have on the resulting data and the trajectory of your research. Therefore, it is crucial to carefully select those that will give you the most adequate answers to your questions.
Understandably, answering the questions is a challenge on its own! In complex areas like cardiovascular or neurological diseases, navigating these challenges becomes even more intricate.
At Ncardia, we recognize the complexities inherent in various research areas and the opportunities brought by iPSC technology. Our iPSC-platform is design to confidently advance your RNA-based therapeutic, empowering you to make more confident decisions.
 Learn more about Ncardia's iPSC-platform to screen RNA therapeutics. Download the poster now
Learn more about Ncardia's iPSC-platform to screen RNA therapeutics. Download the poster now
References
- Fomivirsen approved for CMV retinitis. (1998, October 1). PubMed. https://pubmed.ncbi.nlm.nih.gov/11365956/
- Geary, R. S., Baker, B. F., & Crooke, S. T. (2015). Clinical and Preclinical Pharmacokinetics and Pharmacodynamics of Mipomersen (KynamRo®): A Second-Generation Antisense oligonucleotide inhibitor of apolipoprotein B. Clinical Pharmacokinetics, 54(2), 133–146. https://doi.org/10.1007/s40262-014-0224-4
- Gales, L. (2019). Tegsedi (Inotersen): An Antisense Oligonucleotide Approved for the Treatment of Adult Patients with Hereditary Transthyretin Amyloidosis. Pharmaceuticals, 12(2), 78. https://doi.org/10.3390/ph12020078
- Paik, J., & Duggan, S. T. (2019). Volanesorsen: First global approval. Drugs, 79(12), 1349–1354. https://doi.org/10.1007/s40265-019-01168-z
